2 Downloading NCBI-SRA metadata
2.1 Demo Projects for metadata exploration
In this demonstration, we will delve into sample metadata obtained from various randomly selected public microbiome NCBI BioProjects. This includes information from:
- PRJNA477349: 16S rRNA from bushmeat samples collected from Tanzania Metagenome (Multispecies).
- PRJNA802976: Changes to Gut Microbiota following Systemic Antibiotic Administration in Infants (Multispecies).
- PRJNA322554: The Early Infant Gut Microbiome Varies In Association with a Maternal High-fat Diet (Multispecies).
- PRJNA937707: Microbiome associated with spotting disease in the purple sea urchin (Multispecies).
- PRJNA589182: 16S rDNA gene sequencing of the phyllosphere endophytic bacterial communities colonizing wild Populus trichocarpa Raw sequence reads(Multispecies).
- PRJEB13870: Dysbiosis of gut microbiota contributes to the pathogenesis of hypertension (Multispecies).
- PRJNA208226: Colonization patterns of soil microbial communities in the Atacama Desert (Multispecies).
2.2 Download Methods
Various methods are available for downloading sample metadata from the Sequence Read Archive (SRA) or the European Nucleotide Archive (ENA). Each method provides slightly different information, making it prudent to explore and select the one that best aligns with your specific needs and preferences.
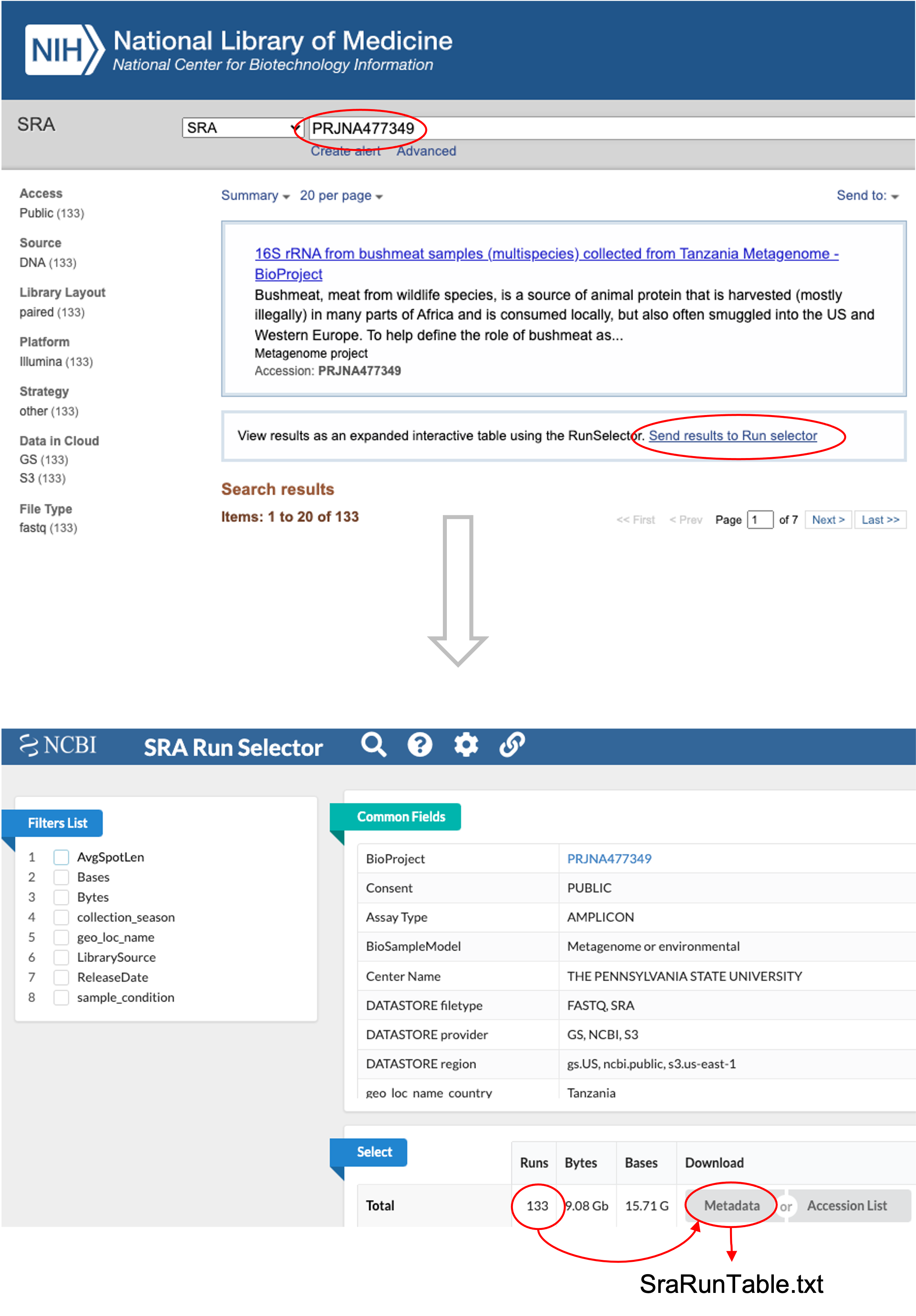
2.2.1 Manually via SRA Run Selector
We can manually retrieve metadata from the SRA database via the SRA Run Selector.
- Note that the SRA filename for metadata is automatically named SraRunTable.txt.
- Users can change the default TXT extension to like CSV if preferred.
- In our demo, we will use CSV to save the metadata file in the
data/folder.
2.2.2 Computationally via Entrez Direct
You can retrieve SRA metadata computationally using Entrez Direct, a set of tools provided by NCBI for accessing the Entrez databases. Here’s a step-by-step guide:
Download Entrez Direct You can download and install
Entrez Directby following the instructions on the NCBI GitHub repository.Add
Entrez Directto PATH After downloading, add theedirectfolder to your system’s PATH environment variable. This allows you to runEntrez Directcommands from any location in your terminal.
Make sure to replace “/path/to/edirect” with the actual path to the edirect folder on your system.
- Download SRA Metadata Use the following Entrez Direct commands to download SRA metadata for specific BioProjects. Replace “PRJ…” with the BioProject accessions of interest.
#!/bin/bash
esearch -db sra -query 'PRJNA477349[bioproject]' | efetch -format runinfo >data/runinfo_PRJNA477349_metadata.csv;
esearch -db sra -query 'PRJNA802976[bioproject]' | efetch -format runinfo >data/runinfo_PRJNA802976_metadata.csv;
esearch -db sra -query 'PRJNA322554[bioproject]' | efetch -format runinfo >data/runinfo_PRJNA322554_metadata.csv;
esearch -db sra -query 'PRJNA937707[bioproject]' | efetch -format runinfo >data/runinfo_PRJNA937707_metadata.csv;
esearch -db sra -query 'PRJNA589182[bioproject]' | efetch -format runinfo >data/runinfo_PRJNA589182_metadata.csv;
esearch -db sra -query 'PRJEB13870[bioproject]' | efetch -format runinfo >data/runinfo_PRJEB13870_metadata.csv;
esearch -db sra -query 'PRJNA208226[bioproject]' | efetch -format runinfo >data/runinfo_PRJNA208226_metadata.csv;
2.2.3 Computationally using pysradb
-
Create pysradb environment
The
pysradbtool can obtain metadata from SRA and ENA. Here we will create an independent environment and install pysradb. We can delete this env when no longer needed. To learn more click here.
- First, we create a
pysradb environmentand install the pysradb tool. - Then we use
pysradbto download the SRA metadata on CLI.
- Download using a
bashscript
#!/bin/bash
# Shell script: workflow/scripts/pysradb_sra_metadata.sh
pysradb metadata PRJNA477349 --detailed >data/PRJNA477349_pysradb.csv
pysradb metadata PRJNA802976 --detailed >data/PRJNA802976_pysradb.csv
pysradb metadata PRJNA322554 --detailed >data/PRJNA322554_pysradb.csv
pysradb metadata PRJNA937707 --detailed >data/PRJNA937707_pysradb.csv
pysradb metadata PRJNA589182 --detailed >data/PRJNA589182_pysradb.csv
pysradb metadata PRJEB13870 --detailed >data/PRJEB13870_pysradb.csv
pysradb metadata PRJNA208226 --detailed >data/PRJNA208226_pysradb.csv- Download using a
pythonscript
# Python script: workflow/scripts/pysradb_sra_metadata.py
import os
import sys
import csv
import pandas as pd
from pysradb.sraweb import SRAweb
db = SRAweb()
df = db.sra_metadata('PRJNA477349', detailed=True)
df.to_csv('data/PRJNA477349_pysradb_metadata.csv', index=False)
db = SRAweb()
df = db.sra_metadata('PRJNA802976', detailed=True)
df.to_csv('data/PRJNA802976_pysradb_metadata.csv', index=False)
db = SRAweb()
df = db.sra_metadata('PRJNA322554', detailed=True)
df.to_csv('data/PRJNA322554_pysradb_metadata.csv', index=False)
db = SRAweb()
df = db.sra_metadata('PRJNA937707', detailed=True)
df.to_csv('data/PRJNA937707_pysradb_metadata.csv', index=False)
db = SRAweb()
df = db.sra_metadata('PRJNA589182', detailed=True)
df.to_csv('data/PRJNA589182_pysradb_metadata.csv', index=False)
db = SRAweb()
df = db.sra_metadata('PRJEB13870', detailed=True)
df.to_csv('data/PRJEB13870_pysradb_metadata.csv', index=False)
db = SRAweb()
df = db.sra_metadata('PRJNA208226', detailed=True)
df.to_csv('data/PRJNA208226_pysradb_metadata.csv', index=False)I sometimes experience ConnectionError when using python method. Try a different method if that happens.
2.3 Filtering user-specified information
Using keywords to search any extensive database helps filter user-specified information, such as certain disease-related studies.
#!/bin/bash
# Install pysradb using mamba package manager
mamba install -c bioconda pysradb
# Search the SRA database for studies related to "Amplicon" and limit to 100 results
pysradb search --db sra -q Amplicon --max 100 > sra_amplicon_studies.csv
# Search the ENA database for studies related to "Amplicon" and limit to 100 results
pysradb search --db ena -q Amplicon --max 100 > ena_amplicon_studies.csv